In this post, we will discuss:
- How to check for missing values
- Different methods to handle missing values
In the example, I demonstrate now there are six columns and seven rows of data.
I imported this data set into python and all the missing values are denoted by NaN (Not-A-Number)
A) Checking for missing values
The following picture shows how to count total number of missing values in entire data set and how to get the count of missing values -column wise.
B) Handling missing values
1) Dropping the missing values
Before deleting the missing values, we should be know the following concept. There are three types of missing values:
- Missing Completely at Random (MCAR)- ignorable
- Missing at Random (MAR) - ignorable
- Missing Not at Random (MNAR) - Not ignorable
To delete/ignore the missing values, it should not be of last type-MNAR. To understand more about these, I request you to read these interesting answers on stackexchange, especially the second answer by Mr. Wayne.
This option should be used when other methods of handling the missing values are not useful. In our example, there was only a one row where there were no single missing values. So only that row was retained when we used dropna() function.
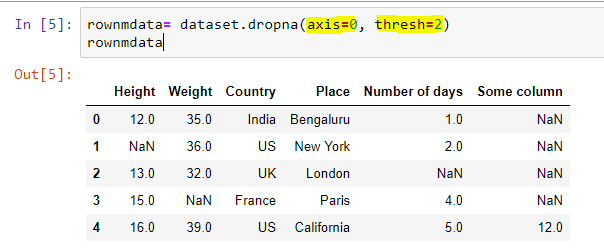
b) Dropping the entire row/column only when there are multiple missing values in the row
As we have seen, the last method of dropping the entire row even when there is only a single missing value is little harsh, we can specify a threshold number of non-missing values before deleting the row. Suppose we want to drop the drop only if there are less than say 2 non-missing values, then we case the following code:
As we have seen, the last method of dropping the entire row even when there is only a single missing value is little harsh, we can specify a threshold number of non-missing values before deleting the row. Suppose we want to drop the drop only if there are less than say 2 non-missing values, then we case the following code:
c) Dropping the entire column
2) Imputing the missing values
a) Replacing with a given value
i) Replacing with a given number, let us say with 0.
ii) Replacing with a string, let us say with 'Mumbai'.
Replacing the missing values with a string could be useful where we want to treat missing values as a separate level.
b) Replacing with mean: It is the common method of imputing missing values. However in presence of outliers, this method may lead to erroneous imputations. In such cases, median is an appropriate measure of central tendency. For some reasons, if you have to use mean values for imputation, then treat the outliers before imputations.
b) Replacing with mean: It is the common method of imputing missing values. However in presence of outliers, this method may lead to erroneous imputations. In such cases, median is an appropriate measure of central tendency. For some reasons, if you have to use mean values for imputation, then treat the outliers before imputations.
c) Replacing with Median: As median is a position based measure of central tendency (middle most item), this method is not affected by presence of outliers.
d) Replacing with Mode:
Mode is the measure of central tendency for nominal scale data.
Replacing with mode is little bit trickier. Because unlike mean and median, mode returns a dataframe. Why? Because if there are two modal values, pandas will show both these values as modes.
For example, let us say our data set is ['A', 'A', 'B', 'C', 'C'].
Here both 'A' and 'C' are the modes as they are repeated equal number of times. Hence mode returns a dataframe containing 'A' and 'C' not a single value.
While replacing with mode, we need to use mode()[0] at the end as shown in the code below.
Mode is the measure of central tendency for nominal scale data.
Replacing with mode is little bit trickier. Because unlike mean and median, mode returns a dataframe. Why? Because if there are two modal values, pandas will show both these values as modes.
For example, let us say our data set is ['A', 'A', 'B', 'C', 'C'].
Here both 'A' and 'C' are the modes as they are repeated equal number of times. Hence mode returns a dataframe containing 'A' and 'C' not a single value.
While replacing with mode, we need to use mode()[0] at the end as shown in the code below.
e) Replacing with previous value - Forward fill
In time series data, replacing with nearby values will be more appropriate than replacing it with mean. Forward fill method fills the missing value with the previous value. For better understanding, I have shown the data column both before and after 'ffill'.
In time series data, replacing with nearby values will be more appropriate than replacing it with mean. Forward fill method fills the missing value with the previous value. For better understanding, I have shown the data column both before and after 'ffill'.
f) Replacing with next value - Backward fill
Backward fill uses the next value to fill the missing value. You can see how it works in the following example.
g) Replacing with average of previous and next value
In time series data, often the average of value of previous and next value will be a better estimate of the missing value. Use the following code to achieve this. I have shown in the following picture how this method works.
h) Interpolation
Similar results can be achieved using interpolation. Interpolation is very flexible with different methods of interpolation such as the default 'linear' (average of ffill and bfill was similar to linear), 'quadratic', 'polynomial' methods (more about this).
Backward fill uses the next value to fill the missing value. You can see how it works in the following example.
g) Replacing with average of previous and next value
In time series data, often the average of value of previous and next value will be a better estimate of the missing value. Use the following code to achieve this. I have shown in the following picture how this method works.
>>> dataset['Number of days'] = pd.concat([dataset['Number of days'].ffill(), dataset['Number of days'].bfill()]).groupby(level=0).mean()
h) Interpolation
Similar results can be achieved using interpolation. Interpolation is very flexible with different methods of interpolation such as the default 'linear' (average of ffill and bfill was similar to linear), 'quadratic', 'polynomial' methods (more about this).
>>> dataset['Number of days']=dataset['Number of days'].interpolate()
i) Model based imputation
We can impute the missing values using model based imputation methods. Popular being imputation using K-nearest neighbors (KNN) (Schmitt et al paper on Comparison of Six Methods for Missing Data Imputation).
KNN is useful in predicting missing values in both continuous and categorical data (we use Hamming distance here)
Even under Nearest neighbor based method, there are 3 approaches and they are given below (Tim's answer on stackechange):
- NN with one single neighbor (1NN)
- with k neighbors without weighting (kNN) or with weighting (wkNN) (Nearest neighbor imputation algorithms: a critical evaluation paper by Beretta and Santaniello)
If you are interested to how to run this KNN based imputation, you can click here for examples in Python and here for R.
You can watch the following video understand better.
Further reading:
https://datascienceplus.com/missing-value-treatment/
Summary
We have seen different methods of handling missing values.
Do you have any questions or suggestions? Feel free to share, I will be happy to interact.







Explained very well with simple examples.........really good work
ReplyDeleteThank you so much for your supportive words.. Glad that this post was useful..
Delete